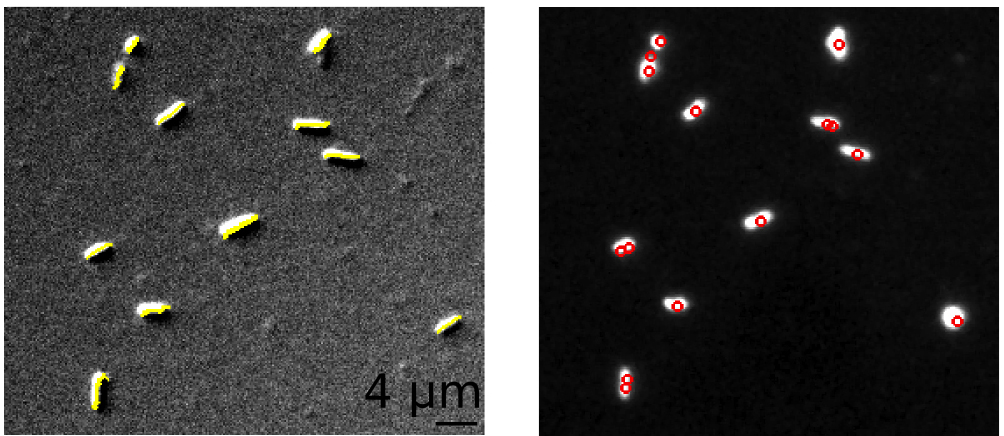
Shown here are images of fixed E. coli K12 (MG1665) cells. For the same set of cells, the algorithm automatically detects cell lengths in DIC images (yellow lines; image on the left) and nucleoids (red circles; image on the right) stained with the fluorescent dye DAPI.
Using the new program, the team then analyzed cell lengths at a population scale in three different strains of bacteria. They found a higher proportion of long cells in bacterial strains which have a defective recombinase A (RecA), an enzyme that repairs breaks in DNA strands and aids recombination of genetic material. However, when cell densities are higher, the proportion of long cells decreases in all strains analyzed. This could mean that cell elongation depends on availability of nutrients. Even more interestingly, Athale and Chaudhari found a linear correlation between number of nucleoids and cell lengths, with length per nucleoid being ~2 micrometres. This suggested that the cells elongate normally but do not form septa, the “lines” along which cells divide. This is the first time a quantitative analysis of this kind has been performed linking cell length and DNA replication in RecA defective strains of bacteria. “This gives us clues on understanding yet another factor that governs cell length variability in a population. It appears that the long cells have turned out so because they managed to elongate but not divide. This could be due to a higher probability of replication fork stalling in RecA deficient strains,” says Athale sharing his excitement on this observation. The study titled “Population length variability and nucleoid numbers in E. coli” is authored by Chaitanya Athale and Hemangi Chaudhari and has appeared in the journal Bioinformatics (27(21):2944-2948). The research was supported by IISER Pune core funds while Hemangi Chaudhari was a KVPY Fellow. -Reported by Shanti Kalipatnapu

